

.
Loading component
This track comprises multiple analyses
Gene annotations from the CSHL gene annotation pipeline developed under NAM project. It is performed through an automated, evidence-based method combining 3rd party software including Mikado, BRAKER and PASA. Further filtered by conservation and AED score, classified into coding and non-coding sets.
Gene annotations from the CSHL gene annotation pipeline developed under NAM project. It is performed through an automated, evidence-based method combining 3rd party software including Mikado, BRAKER and PASA. Further filtered by conservation and AED score, classified into coding and non-coding sets.
Change track style:
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
This track comprises multiple analyses
Gramene maize AGPv4 coding genes mapped with liftoff
Gramene maize AGPv3 coding genes mapped with liftoff
Change track style:
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Track showing sequence in both directions. Only displayed at 1Kb and below.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Manually curated AGPv4 gene models with Apollo Annotation Editor
Change track style:
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Manually curated AGPv4 gene models with Apollo Annotation Editor
Change track style:
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Track showing sequence in both directions. Only displayed at 1Kb and below.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
This track comprises multiple analyses
Gene annotations from the CSHL gene annotation pipeline developed under NAM project. It is performed through an automated, evidence-based method combining 3rd party software including Mikado, BRAKER and PASA. Further filtered by conservation and AED score, classified into coding and non-coding sets.
Gene annotations from the CSHL gene annotation pipeline developed under NAM project. It is performed through an automated, evidence-based method combining 3rd party software including Mikado, BRAKER and PASA. Further filtered by conservation and AED score, classified into coding and non-coding sets.
Change track style:
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
This track comprises multiple analyses
Gramene maize AGPv4 coding genes mapped with liftoff
Gramene maize AGPv3 coding genes mapped with liftoff
Change track style:
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
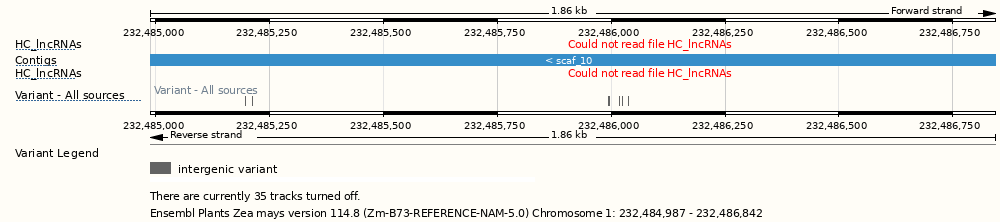
High Confidence lncRNAs
Change track style:
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Track showing underlying assembly contigs.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
High Confidence lncRNAs
Change track style:
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Sequence variants from all sources
Change track style:
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
.






 Off
Off No exon structure without labels
No exon structure without labels No exon structure with labels
No exon structure with labels Expanded without labels
Expanded without labels
 Expanded with labels
Expanded with labels Collapsed without labels
Collapsed without labels Collapsed with labels
Collapsed with labels Coding transcripts only (in coding genes)
Coding transcripts only (in coding genes) Normal
Normal Labels
Labels Structure
Structure Structure with labels
Structure with labels Half height
Half height Stacked
Stacked Stacked unlimited
Stacked unlimited Ungrouped
Ungrouped Wiggle plot
Wiggle plot Normal (collapsed for windows over 200kb)
Normal (collapsed for windows over 200kb)
 Expanded with name (hidden for windows over 10kb)
Expanded with name (hidden for windows over 10kb) Expanded without name
Expanded without name![Follow us on Twitter! [twitter logo]](/i/twitter.png)
