Search
About Brachypodium distachyon
Brachypodium distachyon, like Arabidopsis thaliana, has several features that recommend it as a model plant for functional genomic studies, especially in the grasses. It has a small, diploid genome (~355 Mb), small physical size, a short life-cycle and few growth requirements. Brachypodium is related to the major cereal grain species but is understood to be more closely related to the Triticeae (wheat and barley) than to the other cereals.
Taxonomy ID 15368
Data source Joint Genome Institute
Comparative genomics
What can I find? Homologues, gene trees, and whole genome alignments across multiple species.
 More about comparative analyses
More about comparative analyses
 Phylogenetic overview of gene families
Phylogenetic overview of gene families
 Download alignments (EMF)
Download alignments (EMF)
Variation
What can I find? Short sequence variants.
 More about variation in Ensembl Plants
More about variation in Ensembl Plants
Links
Links (Brachypodium distachyon)
- Gramene species page for Brachypodium
- The International Brachypodium Initiative (IBI) community portal
- The John Innes Centre ModelCrop.org
- MIPS Brachypodium genome database
- Phytozome entry page for Brachypodium distachyon
- Jaiswal Lab at Oregon State University
- INSDC project PRJNA182761
Links (Triticum aestivum)
- MIPS Wheat Genome Database
- ENA study ERP000319: 454 pyrosequencing of the Triticum aestivum (bread wheat) genome to 5X coverage
- ENA study ERP001415: 454 sequencing of Triticum aestivum (bread wheat) cv. Chinese spring cDNA samples from a pool of tissues, from plants under drought stress and from circadian-sampled leaves
- Triticum aestivum ESTs at ENA
- Triticum aestivum UniGene cluster sequences at NCBI




 Convert your data to Brachypodium_distachyon_v3.0 coordinates
Convert your data to Brachypodium_distachyon_v3.0 coordinates Display your data in Ensembl Plants
Display your data in Ensembl Plants

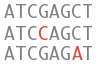

![Follow us on Twitter! [twitter logo]](/i/twitter.png)
