Zm00001e013042
Chromosome 5: 50,084,872-50,091,225 reverse strand.
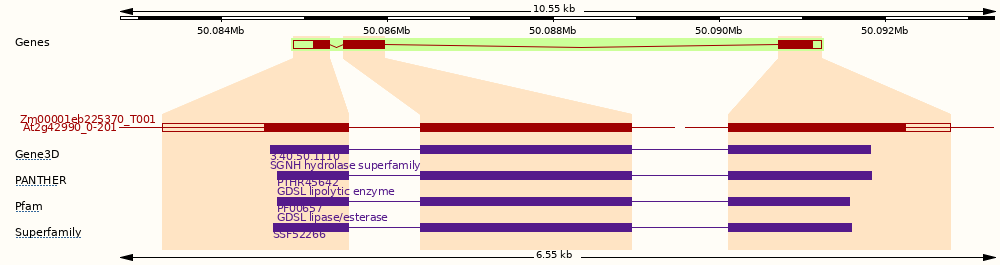
This gene has 1 transcript (splice variant), 354 orthologues and 110 paralogues.
- Name
- Transcript ID
- bp
- Protein
- Translation ID
- Biotype
- UniProt
- RefSeq
- Flags
Zm00001e013042
Chromosome 5: 50,084,872-50,091,225 reverse strand.
This gene has 1 transcript (splice variant), 354 orthologues and 110 paralogues.
.
Protein fingerprints (groups of conserved motifs) from the PRINTS database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Conserved Domain Database models.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the TIGRFAM database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the PIR (Protein Information Resource) Superfamily database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
PANTHER families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the SMART database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the TIGRFAM database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
HAMAP families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
NCBIfam is a collection of protein families based on Hidden Markov Models (HMMs).
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the SMART database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the PROSITE profiles database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the SUPERFAMILY database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
HAMAP families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Structure-Function Linkage Database families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the PROSITE profiles database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the PIR (Protein Information Resource) Superfamily database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein fingerprints (groups of conserved motifs) from the PRINTS database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the PROSITE patterns database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Structure-Function Linkage Database families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
NCBIfam is a collection of protein families based on Hidden Markov Models (HMMs).
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
CATH/Gene3D families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the Pfam database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Conserved Domain Database models.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the PROSITE patterns database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
CATH/Gene3D families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
PANTHER families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the Pfam database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the SUPERFAMILY database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
.